1. Introduction
This protocol allows efficient generation of integration-free iPS cells from a small amount of peripheral blood (<1 ml). Peripheral blood mononuclear cells (PBMCs) are cultured to expand the erythroblast (EB) population. They are then used to derive iPS cells using four recombinant Sendai viral vectors (CytotuneTM, Life Technologies), expressing the four reprogramming factors Oct4, Sox2, Kfl4 and c-Myc.
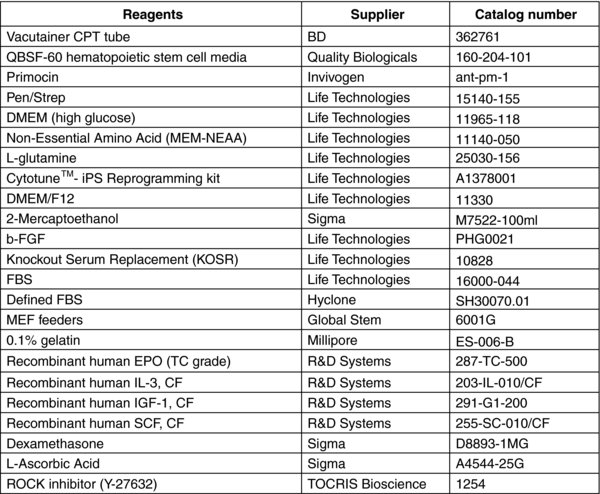
3. Materials and Preparation
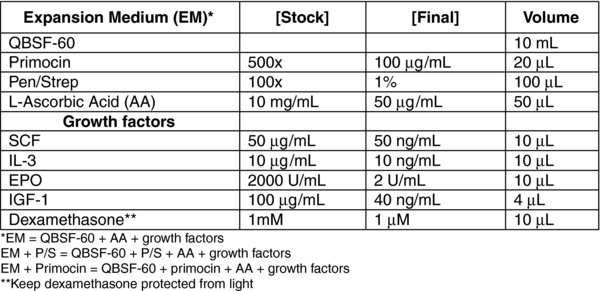
3.1. MEF media (500 ml)
-
DMEM (high glucose): 450 ml
-
FBS: 50 ml
-
MEM-NEAA: 5 ml
-
L-glutamine: 5 ml
-
Pen/Strep: 5 ml
3.2. iPSC Media (500 ml)
-
DMEM/F12: 450 ml
-
Defined FBS: 50 ml
-
MEM-NEAA: 5 ml
-
L-glutamine: 5 ml
-
Pen/strep: 5 ml
-
2-mercaptoethanol: 3.5 μl
-
b-FGF: 10 ng/ml (50 μl of 100 μg/ml stock)
-
L-Ascorbic Acid: 50 μg/ml –add fresh 10 mg/ml stock at each media change
3.3. hESC Media (500 ml)
-
DMEM/F12: 400 ml
-
KOSR: 100 ml
-
MEM-NEAA: 5 ml
-
L-glutamine: 5 ml
-
Pen/strep: 5 ml
-
2-mercaptoethanol: 3.5 μl
-
b-FGF: 10 ng/ml (50 μl of 100 μg/ml stock)
-
L-Ascorbic Acid: 50 μg/ml –add fresh 10 mg/ml stock at each media change
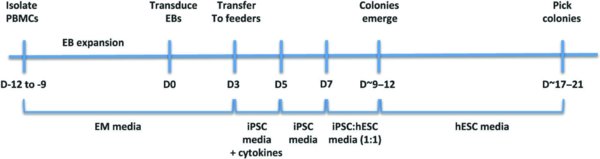
4. Protocol
4.1. D-9 to-12
Collect blood into BD Vacutainer 4 or 8 mL cell preparation tubes (CPT) with sodium citrate or into EDTA or heparinized tubes and Ficoll extract PBMCs. Alternatively, thaw frozen PBMCs.
4.1.1. Fresh cells collected into CPT (8 ml)
-
Draw 8 mL of peripheral blood (PB) into CPT. Invert tube 8–10× and keep upright at room temperature (RT)
-
Centrifuge 30 min at 1,800 RCF at RT (ideally within 2 hrs of collection)
-
Use a sterile transfer pipette to collect buffy coat into sterile 15 mL conical centrifuge tube
-
Bring total volume to 10 mL with sterile 1× PBS, invert several times
-
Centrifuge 15 minutes at 300 RCF and aspirate supernatant
-
Resuspend pellet in 10 mL of sterile 1× PBS and perform cell count (The yield should be ∼1–2×106 cells/ml of PB)
-
Transfer 1–2×106 cells into sterile 15 mL conical centrifuge tube and centrifuge at 300 RCF for 10 min
-
Resuspend pellet in 2 mL of expansion medium (EM) + primocin and transfer to 1 well of a 12-well tissue culture plate
-
Incubate cells at 37°C
-
Centrifuge remaining cells at 300 RCF for 10 min and freeze 1–2×106 cells/vial (Use 90% FBS, 10% DMSO for freezing medium)
4.1.2. Frozen cells
-
11. Thaw 1 vial of PBMCs into 10 mL of QBSF-60 and centrifuge at 300 RCF for 10 min
-
12. Resuspend pellet in 2 mL of EM + primocin and transfer to 1 well of a 12-well plate, incubate at 37°C
4.2. D-6 and D-3 (Pre-Transduction)
Switch media to EM (no antibiotics) at D-6 and collect spent media at D-3 for mycoplasma testing. At D-3, switch back to culturing in EM + P/S.
-
13. Transfer cells to sterile 15 mL conical tube and wash 1× with 1 mL of QBSF-60 to collect non- and loosely adherent cells. Scrape the well with a cell scraper to collect all cells if necessary.
-
14. Centrifuge cells at 300 RCF for 10 min and resuspend in 2 mL of fresh EM + P/S
-
15. Continue to culture in 1 well of a 12-well plate
4.3. D-2-D0 (FACS for Erythroblast markers)
-
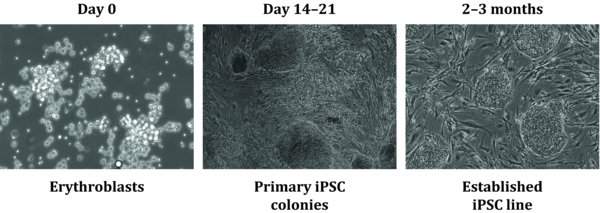
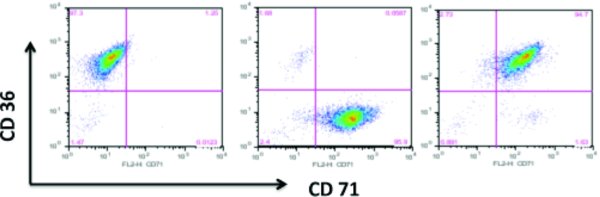
16. EM media expands the erythroblast population from PBMCs. A 2-fold expansion should occur in about 9–12 days with an initial decrease in cell number. When cells are noticeably dividing and have reached the appropriate density, perform FACS to monitor erythroblast expansion using antibodies to erythroblast cell surface markers (see support protocol). When more than 90% of the cells express CD36 and CD71, you can proceed to transduction.
4.4. D0 (Transduction)
4 Sendai viral vectors (CytoTuneTM, Life Technologies) each expressing Oct3/4, Sox2, Klf4, c-Myc are used for transduction. We typically transduce 2.5×105 cells with 10 MOI of each of the four viruses (0.01%-1% efficiency).
-
17. Transfer cells to sterile 15 mL conical tube and wash 1× with 1 mL of QBSF to collect non-adherent and loosely adherent cells
-
18. Count cells
-
19. Centrifuge 2.5×105 cells in 15 mL conical tube and add 1 mL of fresh EM+P/S plus viruses and transfer to one well of a 12 well plate.
-
20. Spinoculation: Centrifuge plate at 2250 rpm at 25°C × 90 min.
-
21. While centrifuging, divide the remaining cells into two tubes, centrifuge, and save one tube for RNA and one for DNA.
-
22. Move centrifuged plate to incubator and maintain at 37°C, 5% CO2, 5% O2, and 90% N2
-
23. Following ∼6–8 hours, add an additional 1 mL of fresh EM + P/S to cells (for a total of 2 ml of EM + P/S)
4.5. D1 (Wash virus)
-
24. Collect and centrifuge cells at 300 RCF in a conical tube for 10 min and resuspend in 2 mL of fresh EM+P/S
4.6. D2 (Plate MEFs)
-
25. Plate MEFs onto 0.1% gelatin coated 6-well TC plates
4.7. D3 (Plate transduced cells)
-
26. Collect cells into 15 mL conical tube and centrifuge at 300 RCF for 10 min.
-
27. Resuspend cells in 6 mL of iPSC media plus growth factors as in EM medium
-
28. Plate 1 mL/well into 6-well MEF plate. Add additional 1.5 mL/well of iPSC media plus growth factors for a total of 2.5 mL/well
-
29. Centrifuge plate at 500 rpm at 25°C × 30 min
4.8. D5–D7
-
30. Feed cells on day 5 with 2.5 mL of iPSC media w/o growth factors
-
31. Feed cells on day 7 with 2.5 mL of iPSC:hESC (1:1) media
-
32. Aspirate and discard floating cells with each feed
4.9. ∼D9–12 (Small colonies emerge)
-
33. Once small colonies appear, feed cells daily with 2 mL of hESC media
-
34. Add additional MEFs as needed (∼1×/wk)
4.10. ∼D13–17 (Cell death)
A significant amount of cell death will occur during this period. Wash wells as needed to remove excess cell debris. Well-defined iPSC colonies will emerge during this period.
4.11. ∼D17–21 (Pick colonies)
-
35. Each colony is picked into one well of a 12-well or 24-well plate pre-coated with MEFs on gelatin in 1 mL/well of hESC media containing 10 μM ROCK inhibitor
-
36. Feed cells in two days, then daily thereafter with 1 mL of hESC media, and continue to expand clones for characterization
Support protocol: FACS analysis of erythroblast surface marker expression
6. Protocol
-
Harvest cells from the 12-well plate into a 15 ml conical tube
-
Centrifuge for 10 min at 400 RCF
-
Discard the supernatant and resuspend cells in 1mL EM media
-
Count cells and transfer 105 cells to round bottom tubes with 3 ml of ice-cold PBS
-
Centrifuge cells for 5 min at 400 RCF
-
Discard the PBS and resuspend cells with 100 μL staining buffer.
-
Stain cells with each of the premade antibody mixtures at 4°C for 30 min: isotype controls (1μl each), CD36 (1μl), CD71 (1μl), CD36 + CD71 (1μl each)
-
Wash cells with 3 ml ice-cold PBS and centrifuge for 10 min at 400 RCF; repeat wash
-
Discard the supernatant by inverting the tube and fix cells with 200 μL 1% paraformaldehyde. Proceed with flow cytometry acquisition.
References
Last revised April 26, 2012. Published December 11, 2012. This chapter should be cited as: Yang, W., Mills, J.A., Sullivan, S., Liu, Y., French, D.L., and Gadue, P., iPSC Reprogramming from human peripheral blood using sendai virus mediated gene transfer (December 11, 2012), StemBook, ed. The Stem Cell Research Community, StemBook, doi/10.3824/stembook.1.73.1, https://www.stembook.org.